My research group is engaged in understanding how macromolecules, both biological and synthetic, assume their sizes and shapes, organize into assemblies, and move around in crowded environments. We employ a combination of theoretical, computational, and experimental techniques to uncover the underlying mechanisms of macromolecular phenomena in Physical Biology and Polymer Physics. The major issues that are under active investigation are the following:
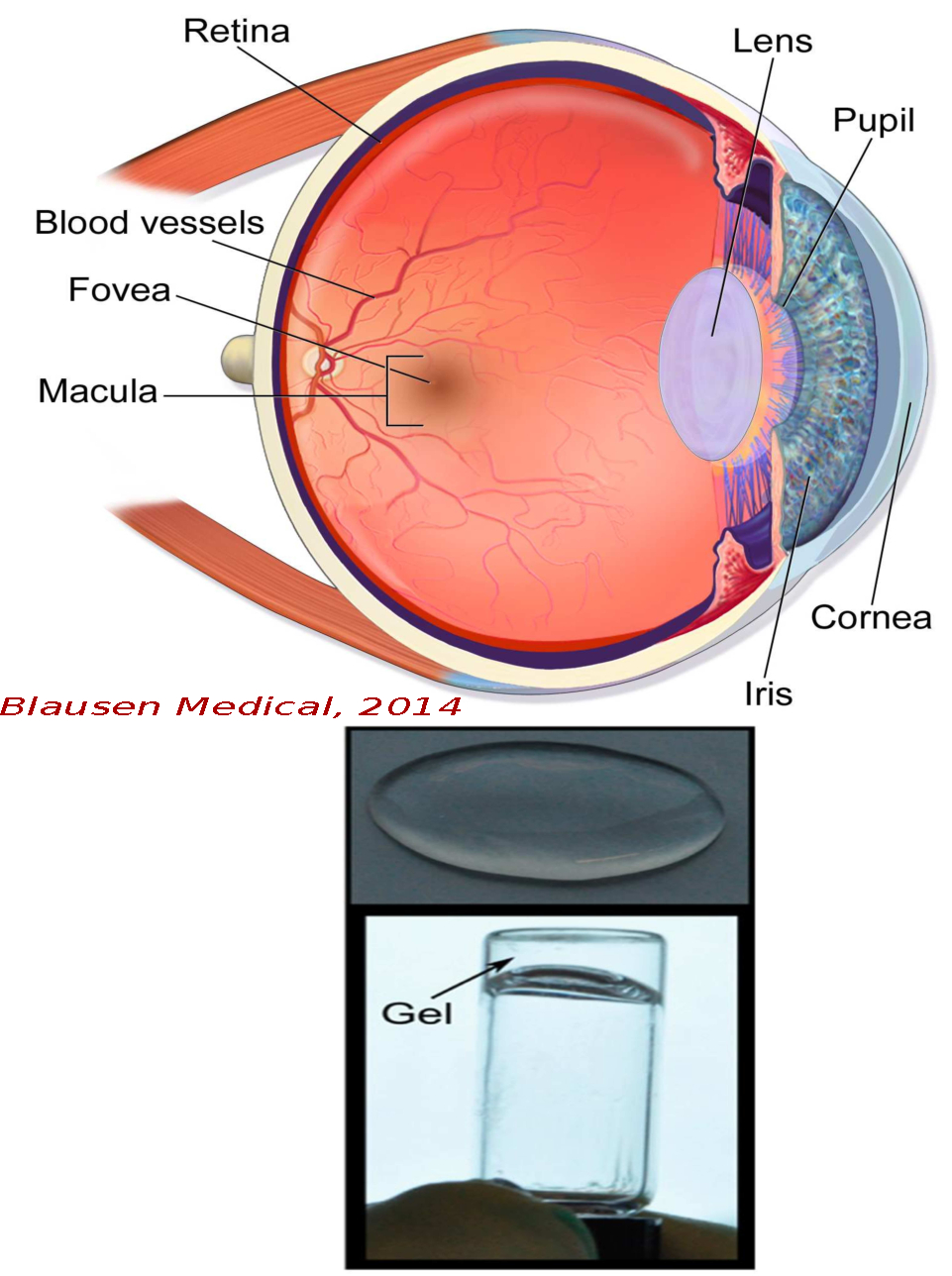
We investigate experimentally the interaction among the various macromolecules (including crystallins, proteoglycans, and collagen) that constitute the major optical components of an eye. The primary objective is to identify the macromolecular mechanisms behind the clumping of proteins in the crystalline lens in order to reverse/remove the onset of cataracts and presbyopia.
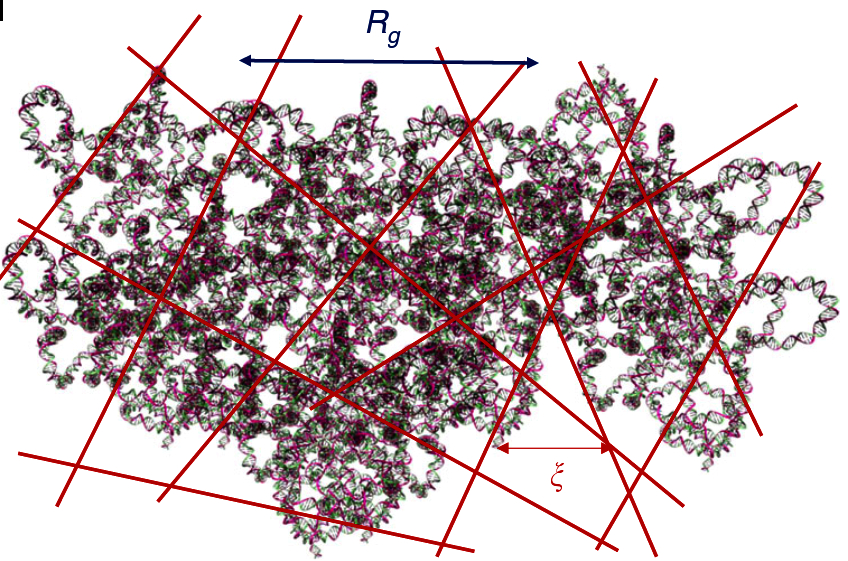
We study gel dynamics using impedence spectroscopy, light scattering and rheology in order to understand counterion dynamics, network elasticity and topological frustration. Our work opens fundamental questions about the behavior of charged macromolecules in crowded environments, in solution conditions of extreme ionic strength, and at high concentrations.
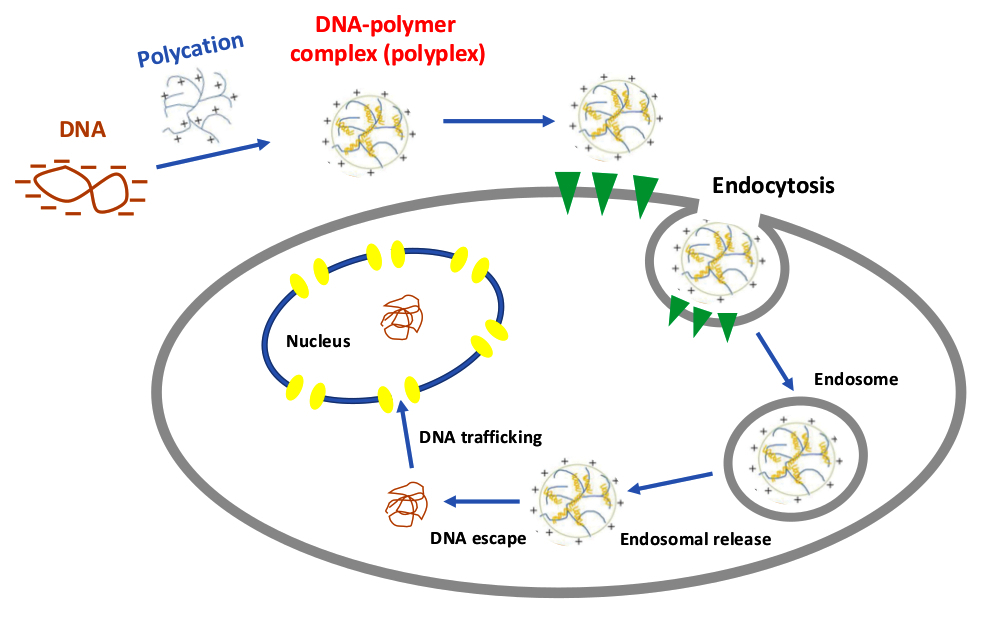
Leber congenital amaurosis (LCA) is a genetic disease associated with mutations in a variety of genes in photoreceptor cells of the retina. In particular, mutations in gene CEP280 lead to defective cilia formation. We employ cationic polymers in order to package nucleic acid material for cell delivery. We are interested in their complexation and uncomplexation kinetics, and factors affecting uptake and release of DNA cargo inside the cell environment.
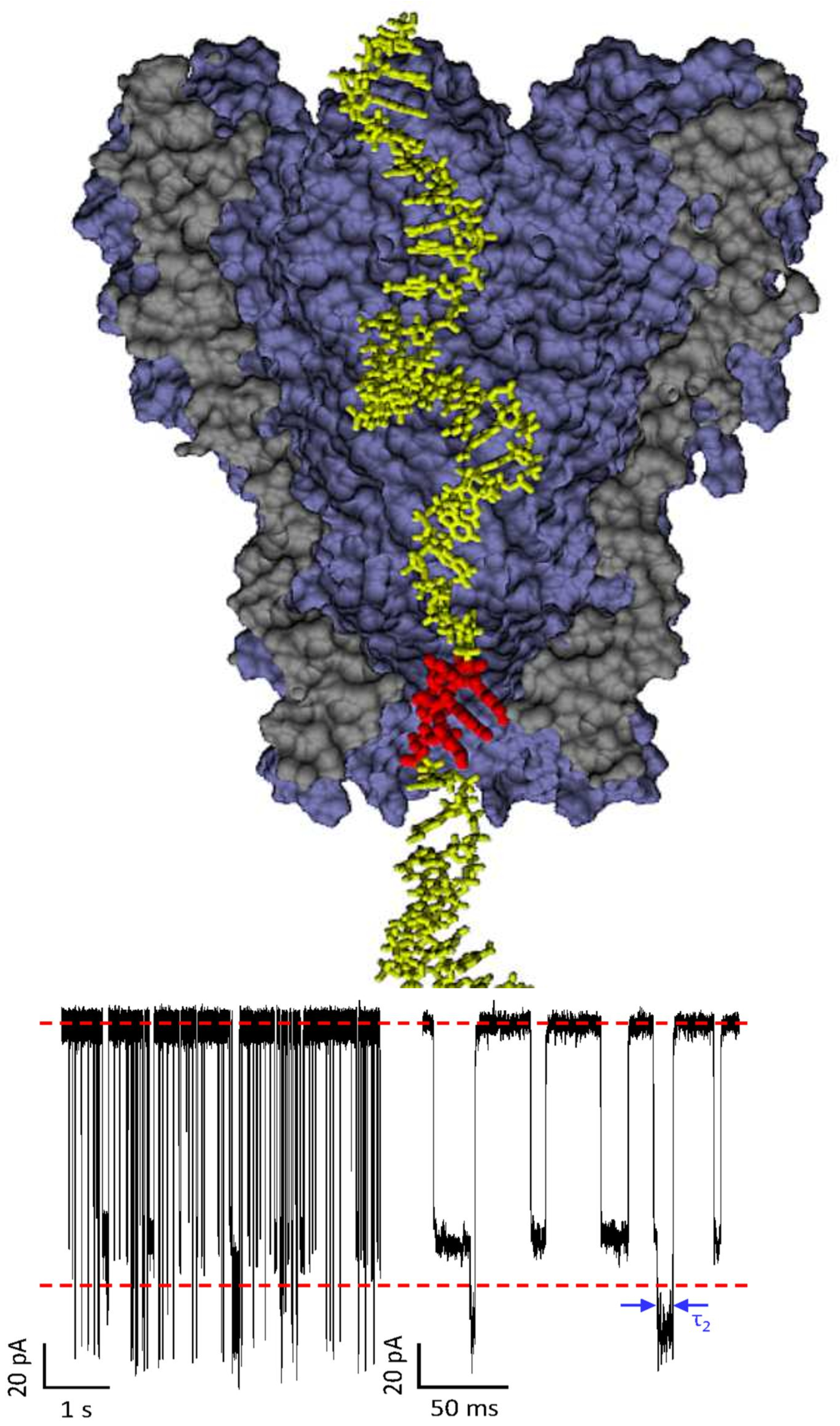
We perform single molecule electrophoresis experiments through a nanopore (biological and synthetic) for translocation of DNA/RNA/proteins and synthetic polyelectrolytes. We also use statistical mechanics, Langevin Dynamics simulations, and the Poisson-Nernst-Plank formalism to elicit stochastic resonance and design experimental strategies for RNA/DNA sequencing to achieve the highest possible accuracy.
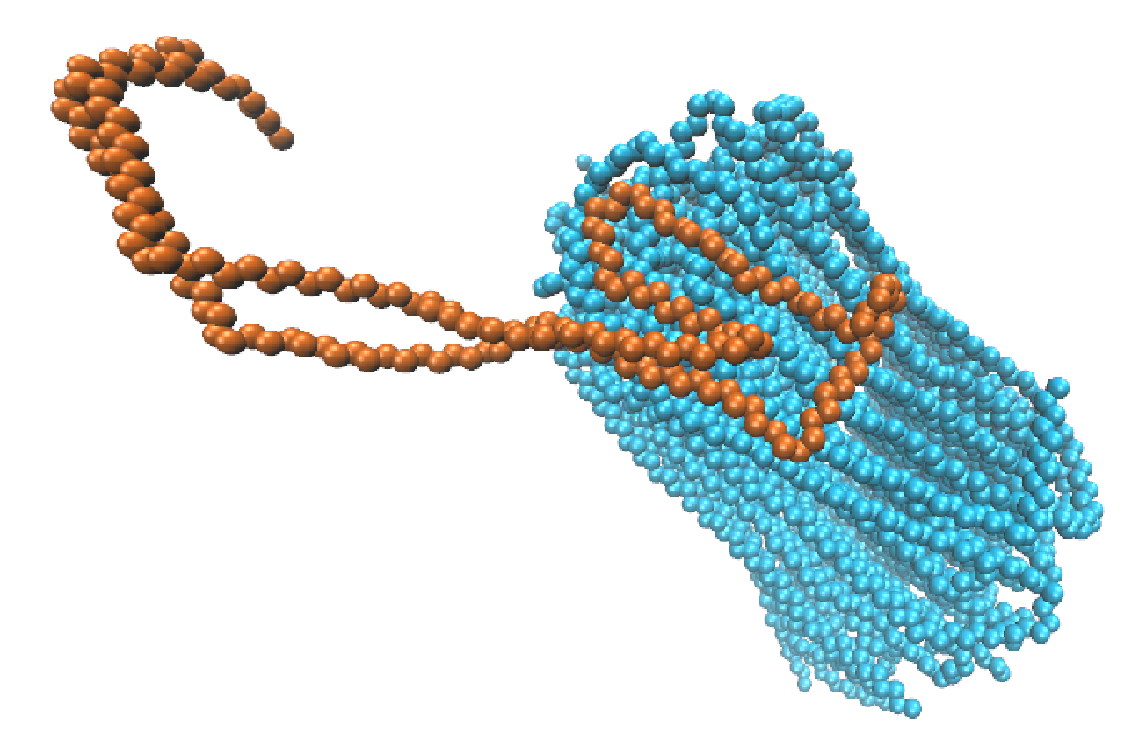
Using Small Angle Light Scattering, SAXS/WAXS, and Differential Dynamic Microscopy, we investigate the crystallization of synthetic polymers and self assembly of biological polymers in efforts to understand the evolution of hierarchical supramolecular structures, memory effects, and storage of energy. These experiments are complemented by computer simulations and statistical mechanics theory.
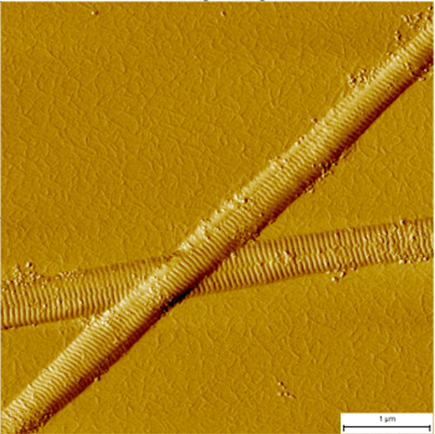
By harnessing our expertise in theory, simulations, and experiments, we investigate the processes by which proteins assemble into large-scale structures. This includes the study of aggregation phenomena, as well as liquid-liquid phase separation in the context of human physiology and disease. Our approach focuses on using fundamental principles to guide thinking in the field and explain findings related to the energetics of signal transduction, in vitro membraneless organelles, and other emerging areas of research.
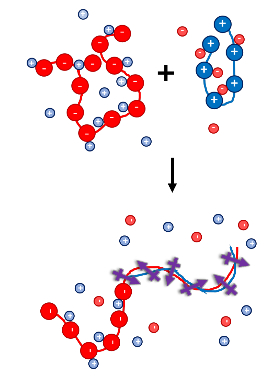
The self-assembly of charged polymers into ordered structures has significant implications in areas including medicine, advanced energy-harnessing devices, and adhesives. We study polyelectrolyte complexes experimentally using dielectric spectroscopy, scattering (SAXS, WAXS, SANS, and SLS/DLS), and microscopy. Our theoretical work focuses on relating this process to the orientation of electric dipole moments in space.